Reporting HLA Antigens for the DRB3/4/5 Locus at the First Field in UNet Would Decrease Rates of Deceased Donor Offer Refusal
(P117) Reporting HLA antigens for the DRB3/4/5 Locus at the first field in UNet would decrease rates of deceased donor offer refusal
Location: Platinum Ballroom
Poster Presenter(s)
Aim: The UNOS organ allocation system, UNet, screens off donors using candidate unacceptable HLA antigens (UAs). For candidates with allelic UAs (e.g. A*02:01) where the low-resolution donor typing includes the unacceptable allele (e.g. A2 or A*02), offers are not screened off by UNet and must be manually reviewed during the virtual crossmatch (VXM). To identify approaches to improve VXM efficiency, we evaluated the potential of donor refusals based on allelic UAs when only low-resolution donor HLA typings are entered.
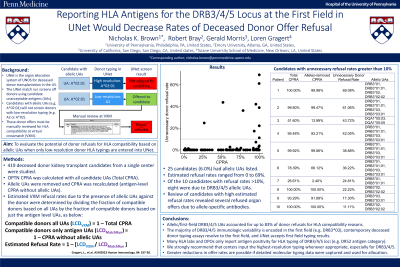
Method: We computed OPTN CPRA, with and without allelic UAs, for 419 candidates listed in UNet for a single center. Estimated VXM refusal rates due to presence of allelic UAs were determined by dividing the fraction of compatible donors based on all UAs by the fraction of compatible donors based on just the antigen-level UAs, and subtracting this number from 1 (Fig. 1).
Results: Twenty-five candidates (6.0%) had allelic UAs. For these candidates, calculated donor refusal rates ranged from 0 to 69%, and the highest refusal rates trended among highly sensitized candidates (Fig. 1). Ten candidates had calculated refusal rates >10%; one patient had allelic UAs to DQA1, one to DRB5, and the other eight to DRB3 (Fig. 2). Overall, DRB3/4/5 allelic UAs accounted for 83% of organ refusals. Review of the candidates uncovered several organ offers that were refused due to allele-specific antibodies, validating the premise.
Conclusion: As donor HLA typing is often reported at a lower resolution than UAs, many incompatible donors are offered to transplant candidates. We calculated the upper boundary of refusal rates due to allelic UAs, and found allelic DRB3/4/5 UAs accounted for up to 83% of donor refusals. Importantly, the majority of DRB3/4/5 immunologic variability is encoded in the first field (e.g. DRB3*03), contemporary deceased donor typing assays resolve to the first field, and UNet accepts first field typing results. However, many HLA labs and OPOs only report locus positivity (e.g. DR52). Reporting the first field for DRB3/4/5 will vastly reduce the rate of positive VXMs, and UNet is currently set up to appropriately screen donors based on intermediate or high-resolution typing results. Therefore, we strongly recommend that centers input the highest-resolution typing whenever appropriate, especially for DRB3/4/5.
Method: We computed OPTN CPRA, with and without allelic UAs, for 419 candidates listed in UNet for a single center. Estimated VXM refusal rates due to presence of allelic UAs were determined by dividing the fraction of compatible donors based on all UAs by the fraction of compatible donors based on just the antigen-level UAs, and subtracting this number from 1 (Fig. 1).
Results: Twenty-five candidates (6.0%) had allelic UAs. For these candidates, calculated donor refusal rates ranged from 0 to 69%, and the highest refusal rates trended among highly sensitized candidates (Fig. 1). Ten candidates had calculated refusal rates >10%; one patient had allelic UAs to DQA1, one to DRB5, and the other eight to DRB3 (Fig. 2). Overall, DRB3/4/5 allelic UAs accounted for 83% of organ refusals. Review of the candidates uncovered several organ offers that were refused due to allele-specific antibodies, validating the premise.
Conclusion: As donor HLA typing is often reported at a lower resolution than UAs, many incompatible donors are offered to transplant candidates. We calculated the upper boundary of refusal rates due to allelic UAs, and found allelic DRB3/4/5 UAs accounted for up to 83% of donor refusals. Importantly, the majority of DRB3/4/5 immunologic variability is encoded in the first field (e.g. DRB3*03), contemporary deceased donor typing assays resolve to the first field, and UNet accepts first field typing results. However, many HLA labs and OPOs only report locus positivity (e.g. DR52). Reporting the first field for DRB3/4/5 will vastly reduce the rate of positive VXMs, and UNet is currently set up to appropriately screen donors based on intermediate or high-resolution typing results. Therefore, we strongly recommend that centers input the highest-resolution typing whenever appropriate, especially for DRB3/4/5.

