Extracting gDNA from Clotted Specimens: Validating the “Crushed Clot”
(P604) Extracting gDNA from clotted specimens
Location: Platinum Ballroom
Poster Presenter(s)
Aim: In efforts to avoid sample rejection, additional invasive procedures to patients, and delays in testing, our laboratory investigated extracting genomic DNA (gDNA) from clotted blood specimens. We assessed the quantity and quality of gDNA from clotted specimens and ensured lack of inhibition in any of our primary PCR-based assays. We also looked to ascertain concordance of testing results obtained from gDNA extracted from clotted blood specimens with results obtained by established acceptable sample sources.
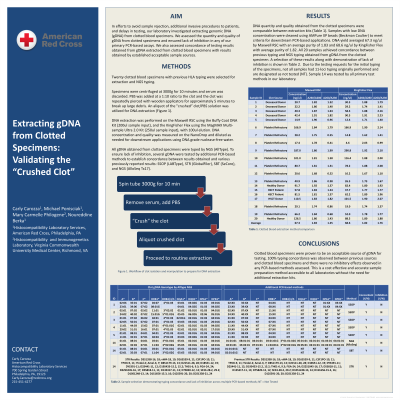
Method: Twenty clotted blood specimens were selected for extraction and NGS typing by AllType. Samples had prior HLA typing from an alternate gDNA source to establish concordance with NGS results of gDNA from the clotted specimens.
Clotted blood specimens were centrifuged and serum discarded. Phosphate buffered saline (PBS) was added at a 1:10 ratio to the clot. The clot was repeatedly pierced with wooden applicators for approximately 5 minutes to break up large debris. An aliquot of 1-2mL of the "crushed” clot/PBS solution was utilized for DNA extraction.
DNA extraction was performed on the Maxwell RSC using the Buffy Coat DNA Kit and the KingFisher Flex using the MagMAX Multi-sample Ultra 2.0 Kit, with 100ul elution. DNA concentration and quality was measured on the NanoDrop and diluted as needed for downstream applications using DNA grade nuclease-free water.
All gDNA obtained from clotted specimens were HLA typed by AllType NGS. To ensure lack of inhibition, several gDNA from clotted specimens were tested by additional PCR-based methods SSOP (LABType), STR (GlobalFiler), SBT (SeCore), and NGS (AlloSeq Tx17) to establish concordance with results obtained to various previously reported results.
Results: DNA quantity and quality of the clotted specimens between extraction kits were found comparable (Table 1) and met criteria for downstream PCR-based applications. DNA yield averaged 67.3 ng/ul by Maxwell RSC with average purity of 1.83 and 68.6 ng/ul by KingFisher Flex with average purity of 1.82. Typing concordance was demonstrated (Table 2) along with lack of inhibition (Table 3) in testing.
Conclusion: Clotted blood specimens were proven to be an acceptable source of gDNA for testing. 100% typing concordance was observed between previous sources and clotted blood specimens. This is a cost effective and accurate sample preparation method accessible to all laboratories without the need for additional extraction kits.
Method: Twenty clotted blood specimens were selected for extraction and NGS typing by AllType. Samples had prior HLA typing from an alternate gDNA source to establish concordance with NGS results of gDNA from the clotted specimens.
Clotted blood specimens were centrifuged and serum discarded. Phosphate buffered saline (PBS) was added at a 1:10 ratio to the clot. The clot was repeatedly pierced with wooden applicators for approximately 5 minutes to break up large debris. An aliquot of 1-2mL of the "crushed” clot/PBS solution was utilized for DNA extraction.
DNA extraction was performed on the Maxwell RSC using the Buffy Coat DNA Kit and the KingFisher Flex using the MagMAX Multi-sample Ultra 2.0 Kit, with 100ul elution. DNA concentration and quality was measured on the NanoDrop and diluted as needed for downstream applications using DNA grade nuclease-free water.
All gDNA obtained from clotted specimens were HLA typed by AllType NGS. To ensure lack of inhibition, several gDNA from clotted specimens were tested by additional PCR-based methods SSOP (LABType), STR (GlobalFiler), SBT (SeCore), and NGS (AlloSeq Tx17) to establish concordance with results obtained to various previously reported results.
Results: DNA quantity and quality of the clotted specimens between extraction kits were found comparable (Table 1) and met criteria for downstream PCR-based applications. DNA yield averaged 67.3 ng/ul by Maxwell RSC with average purity of 1.83 and 68.6 ng/ul by KingFisher Flex with average purity of 1.82. Typing concordance was demonstrated (Table 2) along with lack of inhibition (Table 3) in testing.
Conclusion: Clotted blood specimens were proven to be an acceptable source of gDNA for testing. 100% typing concordance was observed between previous sources and clotted blood specimens. This is a cost effective and accurate sample preparation method accessible to all laboratories without the need for additional extraction kits.

