(P508) Short- and long-read next-generation sequencing approaches to characterize HLA-A*30 null alleles differing in the length of homopolymer C region of exon 4
Location: Platinum Ballroom
Poster Presenter(s)
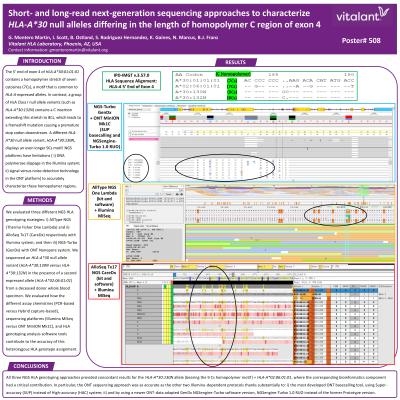
Body: The 5’ end of exon 4 of HLA-A*30:01:01:01 contains a homopolymer stretch of seven cytosines (7Cs), a motif that is common to HLA-A expressed alleles. In contrast, a group of HLA Class I null allele variants (such as HLA-A*30:132N) contains a C insertion extending this stretch to 8Cs, which leads to a frameshift mutation causing a premature stop codon downstream. Moreover, a different HLA-A*30 null allele variant, HLA-A*30:130N, displays an even longer 9Cs motif. It has been postulated that these C insertions were generated from either conversion mechanism with neighboring pseudogenes (such as HLA-J) or slipped-strand mispairing. Next-generation sequencing (NGS) short-read technology from Illumina and long-read technology from Oxford Nanopore (ONT) have revolutionized the field of HLA sequencing. Nevertheless, both approaches have limitations (DNA polymerase slippage in the Illumina system; and signal versus noise detection technology in the ONT platform) to accurately characterize HLA alleles differing in low-complexity sequence regions such as the length of a homopolymer stretch. In this study, we examined the ability of three different NGS HLA genotyping strategies [i) AllType NGS (Thermo Fisher One Lambda) and ii) AlloSeq Tx17 (CareDx) with Illumina system; as well as iii) NGS-Turbo (GenDx) with ONT system] to characterize an HLA-A*30 null allele variant (HLA-A*30:130N versus HLA-A*30:132N) in the presence of a second expressed allele (HLA-A*02:06:01:01) from a deceased donor whole blood sample. Specifically, we evaluated how the different assay chemistries (PCR-based versus Hybrid capture-based), sequencing platforms (Illumina MiSeq versus ONT MinION Mk1C), and HLA genotyping analysis software tools contribute to the accuracy of this HLA genotype assignment.
Conclusion: All three NGS HLA genotyping approaches provided concordant results for the HLA-A*30:130N allele, where the corresponding bioinformatics component had a critical contribution. In particular, the ONT sequencing approach was as accurate as the other two Illumina-dependent protocols thanks substantially to the most developed ONT basecalling tool (using Super-accuracy (SUP) instead of High-accuracy (HAC) system) and by using a newer ONT-data adapted GenDx NGSengine-Turbo software version (NGSengine-Turbo 1.0 RUO instead of the Prototype version).
Conclusion: All three NGS HLA genotyping approaches provided concordant results for the HLA-A*30:130N allele, where the corresponding bioinformatics component had a critical contribution. In particular, the ONT sequencing approach was as accurate as the other two Illumina-dependent protocols thanks substantially to the most developed ONT basecalling tool (using Super-accuracy (SUP) instead of High-accuracy (HAC) system) and by using a newer ONT-data adapted GenDx NGSengine-Turbo software version (NGSengine-Turbo 1.0 RUO instead of the Prototype version).

