(P109) High-throughput, high-resolution HLA typing using NGS-Pronto nanopore sequencing – a multicentre performance study
Location: Platinum Ballroom
Poster Presenter(s)
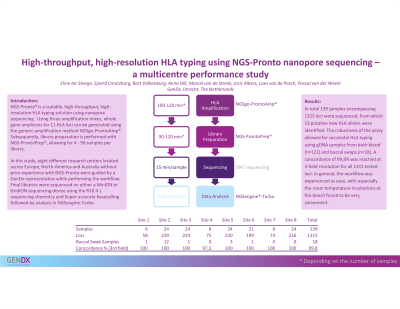
Aim: NGS-Pronto is a scalable high-throughput, high-resolution HLA typing solution using nanopore sequencing. Using three amplification mixes, whole gene amplicons for 11 HLA loci can be generated using NGSgo-ProntoAmp. Subsequently, library preparation is performed with NGS-ProntoPrep, allowing for 4- 96 samples per library. Through this flexibility, NGS-Pronto can be applied in labs with scalable sample turnover. Data analysis is performed with NGSengine®-Turbo software (GenDx) optimized for ONT data analysis. Depending on the number of samples included, the turnaround times, including sequencing and data analysis, take up 5 (4 samples) to 36 hours (96 samples).
Method: To test the performance and ease of use of NGS-Pronto, we tested NGS-Pronto at five different centers spread across Europe, North America and Australia. All centers never used NGS-Pronto before and were guided by a GenDx representative while performing the workflow.
Results: Varying number of samples ranging from 6-24 were tested at the sites. In total, 86 samples and 853 loci were sequenced, from which 11 putative new HLA alleles were identified. The assay was robust enough to allow for successful HLA typing using gDNA samples from buccal swaps (n=17, 20%) of the samples were gDNA samples extracted from buccal swabs. At four sites, 100% concordance was reached at 3-field resolution. One site obtained a concordance of 97%. Here, two samples, originating from family members, contained a deletion of an adenine in exon 6 of HLA-C that was not reported by the NGSengine-Turbo software due to the deletion being present in a homopolymer stretch of four adenines. In general, the workflow is experienced to be easy and especially the room temperature incubations at the bench are found to be very convenient.
Conclusion: The NGS-Pronto workflow is reported to be of easy use, allowing for robust high-throughput third field resolution HLA typing at five centers.
Method: To test the performance and ease of use of NGS-Pronto, we tested NGS-Pronto at five different centers spread across Europe, North America and Australia. All centers never used NGS-Pronto before and were guided by a GenDx representative while performing the workflow.
Results: Varying number of samples ranging from 6-24 were tested at the sites. In total, 86 samples and 853 loci were sequenced, from which 11 putative new HLA alleles were identified. The assay was robust enough to allow for successful HLA typing using gDNA samples from buccal swaps (n=17, 20%) of the samples were gDNA samples extracted from buccal swabs. At four sites, 100% concordance was reached at 3-field resolution. One site obtained a concordance of 97%. Here, two samples, originating from family members, contained a deletion of an adenine in exon 6 of HLA-C that was not reported by the NGSengine-Turbo software due to the deletion being present in a homopolymer stretch of four adenines. In general, the workflow is experienced to be easy and especially the room temperature incubations at the bench are found to be very convenient.
Conclusion: The NGS-Pronto workflow is reported to be of easy use, allowing for robust high-throughput third field resolution HLA typing at five centers.

