(P110) Sensitive and fast NGS-based chimerism monitoring using ONT sequencing
Location: Platinum Ballroom
Poster Presenter(s)
Aim: Chimerism monitoring after hematopoietic stem cell transplantation enables prediction of an upcoming relapse. Enhanced sensitivity of chimerism assays allows for earlier detection, thereby increasing the time available for treatment. NGStrack (GenDx) is an NGS-based chimerism monitoring assay for research use only, using 34 bi-allelic indel markers and Illumina sequencing. Illumina sequencing is robust, but relatively slow and requires a large equipment investment. Oxford Nanopore Technologies (ONT) sequencing combines rapid analysis and scalability with a small footprint and low investment requirements. Here, we extrapolate the design of NGStrack to be compatible with ONT sequencing.
Method: Artificial chimerism samples were created by diluting (cell line) DNA in DNA, resulting in 9 series, each consisting of 16 chimeric samples. NGStrack primers were used for amplification of the 34 markers, followed by indexing using ONT specific indices, sample pooling and custom ONT library preparation. Libraries were sequenced on a MinION flow cell (R10.4.1) on a MinION Mk1B or GridION. Basecalling was performed real-time with MinKNOW Super-accurate (SUP) basecalling algorithm. Data was analyzed in TRKengine 1.4 (GenDx) using optimized settings for ONT data.
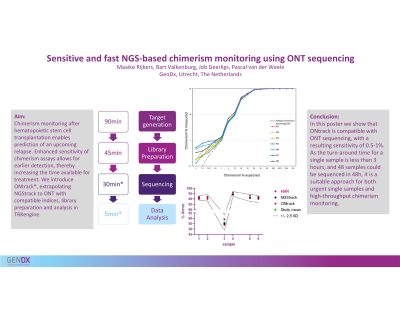
Results: Generating ~200.000 reads per sample, chimerism levels as low as 0.5-1% could be accurately measured, approaching the sensitivity of NGStrack using Illumina (up to 0.1%). Despite increased sequencing noise with ONT compared to Illumina, , which may be a limiting factor for sensitivity, an overall linearity of R2=0.999 was obtained upon analysis. In 2.5 hours, enough reads were generated to genotype 96 samples. When sequencing for 24 hours up to 48 samples can be monitored. For a single monitoring sample, sufficient reads can be obtained within 35 min of sequencing, resulting in a single sample turnaround time of less than 3 hours from DNA to results.
Conclusion: Here we show that NGStrack is compatible with ONT sequencing, with a resulting sensitivity of 0.5-1%. As the turn-around time for a single sample is less than 3 hours, and 48 samples could be sequenced in 48h, it is a suitable approach for both urgent single samples and high-throughput chimerism monitoring.
Method: Artificial chimerism samples were created by diluting (cell line) DNA in DNA, resulting in 9 series, each consisting of 16 chimeric samples. NGStrack primers were used for amplification of the 34 markers, followed by indexing using ONT specific indices, sample pooling and custom ONT library preparation. Libraries were sequenced on a MinION flow cell (R10.4.1) on a MinION Mk1B or GridION. Basecalling was performed real-time with MinKNOW Super-accurate (SUP) basecalling algorithm. Data was analyzed in TRKengine 1.4 (GenDx) using optimized settings for ONT data.
Results: Generating ~200.000 reads per sample, chimerism levels as low as 0.5-1% could be accurately measured, approaching the sensitivity of NGStrack using Illumina (up to 0.1%). Despite increased sequencing noise with ONT compared to Illumina, , which may be a limiting factor for sensitivity, an overall linearity of R2=0.999 was obtained upon analysis. In 2.5 hours, enough reads were generated to genotype 96 samples. When sequencing for 24 hours up to 48 samples can be monitored. For a single monitoring sample, sufficient reads can be obtained within 35 min of sequencing, resulting in a single sample turnaround time of less than 3 hours from DNA to results.
Conclusion: Here we show that NGStrack is compatible with ONT sequencing, with a resulting sensitivity of 0.5-1%. As the turn-around time for a single sample is less than 3 hours, and 48 samples could be sequenced in 48h, it is a suitable approach for both urgent single samples and high-throughput chimerism monitoring.

