(P105) An HLA genotype-informed dPCR assay for monitoring donor-derived cfDNA
Location: Platinum Ballroom
- BL
Poster Presenter(s)
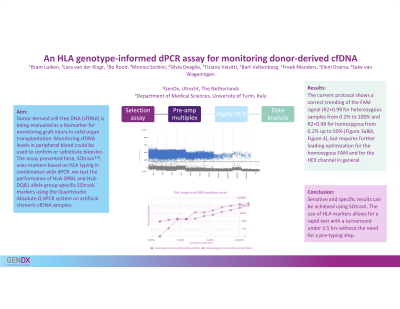
Aim: Donor-derived cell-free DNA (cfDNA) is widely being evaluated as a biomarker for monitoring graft injury and/or rejection in solid organ transplantation (SOT). Monitoring cfDNA levels in peripheral blood could be used to confirm and even substitute invasive, expensive and subjective biopsies. Methods should operate using low quantities of cfDNA and be sensitive, scalable and fast. Digital PCR (dPCR) meets these requirements and the assay presented here eliminates the need for donor pre-transplant DNA by using informative markers based on known data, the HLA typing. Here we test the performance of HLA-DRB1 and HLA-DQB1 allele group-specific markers using the Absolute Q dPCR system, applying a microfluidic array. For application in sex-mismatched transplant, a gender-specific marker is also included.
Method: To test assay performance, gDNA from various cell lines was mixed to create artificial chimeric samples, ranging from high to low chimerism percentages. These mixtures were sheared to cfDNA size. Multiplexed pre-amplification was performed specific for all DRB1 markers, for all DQB1 markers or for the gender. Subsequently a 2-color allele group-specific multiplexed reaction was performed on the Absolute Q dPCR targeting only the informative markers. One color represents the ‘donor’ HLA and a second color the ‘recipient’ HLA allele group. Pre-amplified samples were loaded onto MAP16 plates together with MasterMix and buffer and followed by amplification. Absolute Q software and Excel were used for data analysis.
Results: The dPCR assay required several optimization steps to allow for analysis. These included PCR optimization (optimization of pre-PCR cycle number, dPCR input volumes and cycling temperatures) as well as and optimization of loading the machine to prevent array overloading. Results of the optimized protocol demonstrated that the new assay based on HLA genotype allowed for a linear performance of the assay over a wide range of artificial cfDNA chimeric samples. Turn-around-time of the assay was within 2,5 hrs for 16 samples, with 2 manual steps.
Conclusion: The cfDNA prototype assay for Absolute Q dPCR shows sensitive and specific results. The use of HLA and gender-specific markers allow for a rapid test with a turnaround under 2,5 hours without the need for a pre-typing step.
Method: To test assay performance, gDNA from various cell lines was mixed to create artificial chimeric samples, ranging from high to low chimerism percentages. These mixtures were sheared to cfDNA size. Multiplexed pre-amplification was performed specific for all DRB1 markers, for all DQB1 markers or for the gender. Subsequently a 2-color allele group-specific multiplexed reaction was performed on the Absolute Q dPCR targeting only the informative markers. One color represents the ‘donor’ HLA and a second color the ‘recipient’ HLA allele group. Pre-amplified samples were loaded onto MAP16 plates together with MasterMix and buffer and followed by amplification. Absolute Q software and Excel were used for data analysis.
Results: The dPCR assay required several optimization steps to allow for analysis. These included PCR optimization (optimization of pre-PCR cycle number, dPCR input volumes and cycling temperatures) as well as and optimization of loading the machine to prevent array overloading. Results of the optimized protocol demonstrated that the new assay based on HLA genotype allowed for a linear performance of the assay over a wide range of artificial cfDNA chimeric samples. Turn-around-time of the assay was within 2,5 hrs for 16 samples, with 2 manual steps.
Conclusion: The cfDNA prototype assay for Absolute Q dPCR shows sensitive and specific results. The use of HLA and gender-specific markers allow for a rapid test with a turnaround under 2,5 hours without the need for a pre-typing step.
